The emergence of Complex AI Systems
because of the potential threat but also because of how even if AI doesn't kill us it definitely already is making most of us in the developed world symbiotic with AI more and more. However, I'm thinking that I will basically intersperse these articles more at random along the way so that you won't be just overwhelmed with them.
The whole point of this is for more and more people to begin to understand just how much change in a short period of time AI is going to be bringing to the world in this century. It is my hope that we can limit the chaos and death of humans from AI and increase it's effectiveness and helpfulness.
For example, at one point when I was 18 I believed that Each person should own their own personal robot that could go to work and do menial jobs for them so they could stay home and be creative artists, explorers, musicians in whatever positive creative direction they wanted to go. For now, only those who have financial backing can be like this or who work 4 hours and create 4 hours on a more daily basis of every day Work Play in their lives. At times even as a businessman I tried to live this life sometimes successfully so I could play with my kids, earn a living and travel at the same time. But, I never had a robot to "go to work for me". However, I believe my blog now can help me do both for those of you who are evolved enough in thought and enlightenment to benefit from what I'm writing and compiling about.
The following chart will be my research names I will drill down so you can better understand (for better and for worse) AI. Since Evo-Devo is my first I will concentrate for now on "Evolution and Adaptation" for the first group of articles.
- ... and developmental biology began, with the ... evolutionary biology to form evo- devo.
- Evo-devo by the Understanding Evolution team. Development is the process ... Studies of development are important to evolutionary biology for several reasons.
- Apr 25, 2000 ... The official union occurred in 1999 when evolutionary developmental biology, or “evo-devo,” was granted its own division in the Society for ...
- EvoDevo publishes articles on a broad range of topics associated with the ... budding, or regeneration are amongst the greatest challenges in biology.
- Oct 26, 2009 ... Cliff Tabin: Evolutionary developmental biology, or "evo devo," is a broad term that encompasses a lot of things. And different people use the ...
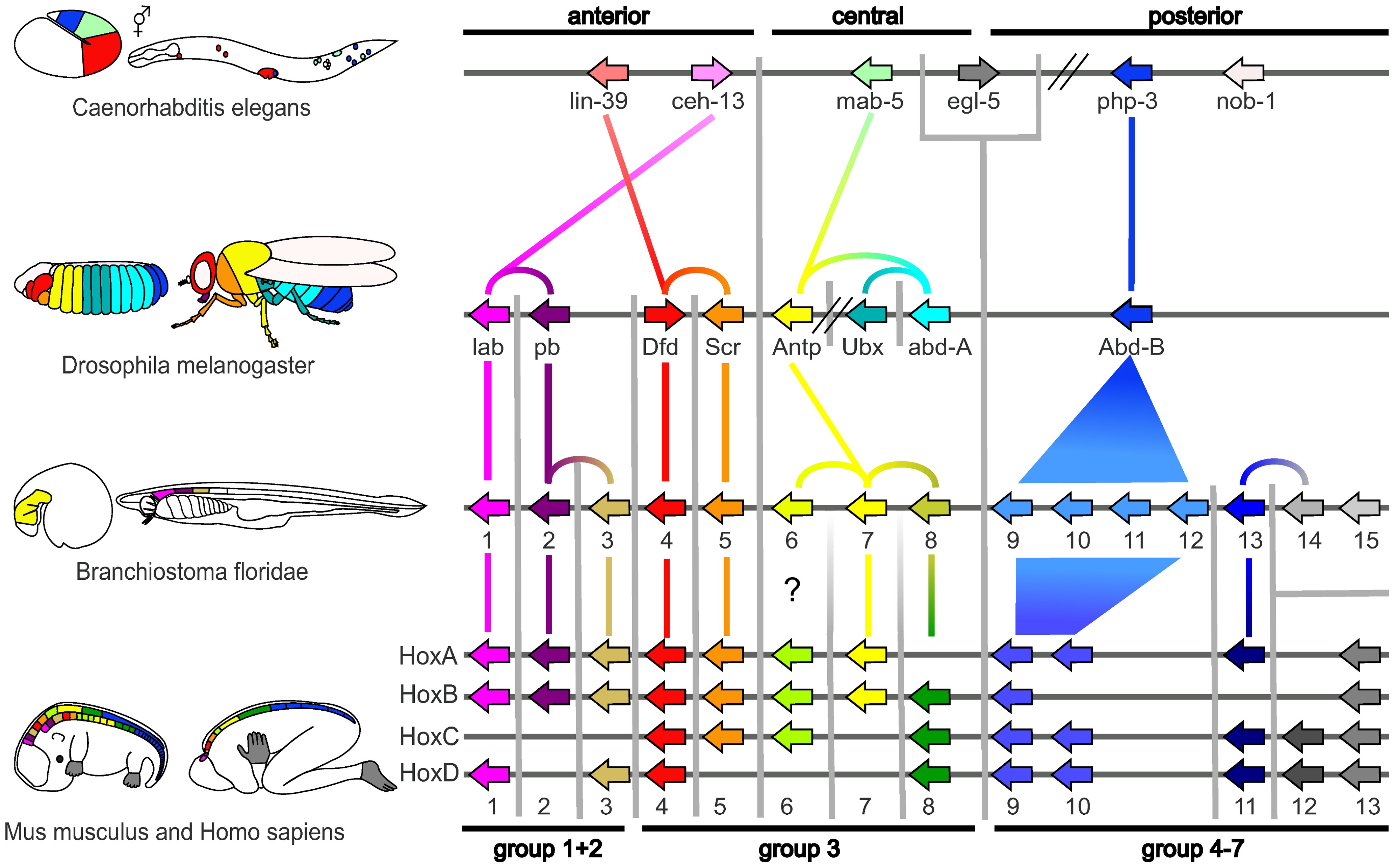
Homologous hox genes in such different animals as insects and vertebrates control embryonic development and hence the form of adult bodies. This shows that these genes have been highly conserved through hundreds of millions of years of evolution.
Evolutionary developmental biology (informally, evo-devo) is a field of biological research that compares the developmental processes of different organisms to infer the ancestral relationships between them and how developmental processes evolved.Part of a series on Evolutionary biology 
The field grew from 19th century beginnings, where embryology faced a mystery: zoologists did not know how embryonic development was controlled at the molecular level. Charles Darwin noted that having similar embryos implied common ancestry, but little progress was made until the 1970s. Then, recombinant DNA technology at last brought embryology together with molecular genetics. A key early discovery was of homeotic genes that regulate development in a wide range of eukaryotes.
The field is characterised by some key concepts, which took biologists by surprise. One is deep homology, the finding that dissimilar organs such as the eyes of insects, vertebrates and cephalopod molluscs, long thought to have evolved separately, are controlled by similar genes such as pax-6, from the evo-devo gene toolkit. These genes are ancient, being highly conserved among phyla; they generate the patterns in time and space which shape the embryo, and ultimately form the body plan of the organism. Another is that species do not differ much in their structural genes, such as those coding for enzymes; what does differ is the way that gene expression is regulated by the toolkit genes. These genes are reused, unchanged, many times in different parts of the embryo and at different stages of development, forming a complex cascade of control, switching other regulatory genes as well as structural genes on and off in a precise pattern. This multiple pleiotropic reuse explains why these genes are highly conserved, as any change would have many adverse consequences which natural selection would oppose.
New morphological features and ultimately new species are produced by variations in the toolkit, either when genes are expressed in a new pattern, or when toolkit genes acquire additional functions. Another possibility is the Neo-Lamarckian theory that epigenetic changes are later consolidated at gene level, something that may have been important early in the history of multicellular life.
Contents
History
Ernst Haeckel argued for recapitulation of evolutionary development in the embryo, using embryo drawings to emphasise the genuine similarities.[1]Further information: History of evolutionary thoughtRecapitulation
Main article: Recapitulation theoryA recapitulation theory of evolutionary development was proposed by Étienne Serres in 1824–26, echoing the 1808 ideas of Johann Friedrich Meckel, which in turn had been hinted at by Aristotle. They argued that the embryos of 'higher' animals went through or recapitulated a series of stages, each of which resembled an animal lower down the great chain of being. For example, the brain of a human embryo looked first like that of a fish, then in turn like that of a reptile, bird, and mammal before becoming clearly human. The embryologist Karl Ernst von Baer opposed this, arguing in 1828 that there was no linear sequence as in the great chain of being, based on a single body plan. Von Baer instead recognised four distinct animal body plans: radiate, like starfish; molluscan, like clams; articulate, like lobsters; and vertebrate, like fish. Zoologists then largely abandoned recapitulation, though Ernst Haeckel revived it in 1866.[2][3][4][5][6]
Evolutionary morphology
A. Lancelet (a chordate), B. Larval tunicate, C. Adult tunicate. Kowalevsky saw that the notochord (1) and gill slit (5) are shared by tunicates and vertebrates.Further information: Morphology (biology) and Body planFrom the early 19th century through most of the 20th century, embryology faced a mystery. Animals were seen to develop into adults of widely differing body plan, often through similar stages, from the egg, but zoologists knew almost nothing about how embryonic development was controlled at the molecular level, and therefore equally little about how developmental processes had evolved.[7] Charles Darwin argued that a shared embryonic structure implied a common ancestor. As an example of this, Darwin cited in his 1859 book On the Origin of Species the shrimp-like larva of the barnacle, whose sessile adults looked nothing like other arthropods; Linnaeus and Cuvier had classified them as molluscs.[8][9] Darwin also noted Alexander Kowalevsky's finding that the tunicate, too, was not a mollusc, but in its larval stage had a notochord and pharyngeal slits which developed from the same germ layers as the equivalent structures in vertebrates, and should therefore be grouped with them as chordates.[8][10] 19th century zoology thus converted embryology into an evolutionary science, connecting phylogeny with homologies between the germ layers of embryos. Zoologists including Fritz Müller proposed the use of embryology to discover phylogenetic relationships between taxa. Müller demonstrated that crustaceans shared the Nauplius larva, identifying several parasitic species that had not been recognised as crustaceans. Müller also recognised that natural selection must act on larvae, just as it does on adults, giving the lie to recapitulation, which would require larval forms to be shielded from natural selection.[8] Two of Haeckel's other ideas about the evolution of development have fared better than recapitulation: he argued in the 1870s that changes in the timing (heterochrony) and changes in the positioning within the body (heterotopy) of aspects of embryonic development would drive evolution by changing the shape of a descendant's body compared to an ancestor's. It took a century before these ideas were shown to be correct.[11][12][13] In 1917, D'Arcy Thompson wrote a book on the shapes of animals, showing with simple mathematics how small changes to parameters, such as the angles of a gastropod's spiral shell, can radically alter an animal's form, though he preferred mechanical to evolutionary explanation.[14][15] But for the next century, without molecular evidence, progress stalled.[8]
The modern evolutionary synthesis of the 1930s
Main article: modern evolutionary synthesisIn the so-called modern evolutionary synthesis of the 1930s and 1940s, Ronald Fisher brought together Darwin's theory of evolution, with its insistence on natural selection, heredity, and variation, and Gregor Mendel's laws of genetics into a coherent structure for evolutionary biology. Biologists assumed that an organism was a straightforward reflection of its component genes: the genes coded for proteins, which built the organism's body. Biochemical pathways (and, they supposed, new species) evolved through mutations in these genes. It was a simple, clear and nearly comprehensive picture: but it did not explain embryology.[8][16]
The lac operon
The lac operon. Top:Repressed, Bottom:Active
1: RNA Polymerase, 2: Repressor, 3: Promoter, 4: Operator, 5: Lactose, 6–8: protein-encoding genes, controlled by the switch, that cause lactose to be digestedMain article: lac operonIn 1961, Jacques Monod, Jean-Pierre Changeux and François Jacob discovered the so-called 'lac operon' in the bacterium Escherichia coli. It was a cluster of genes, arranged in a feedback control loop so that its products would only be made when "switched on" by an environmental stimulus. One of these products was an enzyme that splits a sugar, lactose; and lactose itself was the stimulus that switched the genes on. This was a revelation, as it showed for the first time that genes, even in an organism as small as a bacterium, were subject to fine-grained control. The implication was that many other genes were also elaborately regulated.[17]
The birth of evo-devo and a second synthesis
In 1977, a revolution in thinking about evolution and developmental biology began, with the arrival of recombinant DNA technology in genetics, and the papers Ontogeny and Phylogeny by Stephen J. Gould and Evolution by Tinkering by François Jacob. Gould laid to rest Haeckel's interpretation of evolutionary embryology, while Jacob set out an alternative theory.[8] This led to a second synthesis,[18][19] at last including embryology as well as molecular genetics, phylogeny, and evolutionary biology to form evo-devo.[20][21] In 1978, Edward B. Lewis discovered homeotic genes that regulate embryonic development in Drosophila fruit flies, which like all insects are arthropods, one of the major phyla of invertebrate animals.[22] Bill McGinnis quickly discovered homeotic gene sequences, homeoboxes, in animals in other phyla, in vertebrates such as frogs, birds, and mammals; they were later also found in fungi such as yeasts, and in plants.[23][24] There were evidently strong similarities in the genes that controlled development across all the eukaryotes.[25] In 1980, Christiane Nüsslein-Volhard and Eric Wieschaus described gap genes which help to create the segmentation pattern in fruit fly embryos;[26][27] they and Lewis won a Nobel Prize for their work in 1995.[23][28]
Later, more specific similarities were discovered: for example, the Distal-less gene was found in 1989 to be involved in the development of appendages or limbs in fruit flies,[29] the fins of fish, the wings of chickens, the parapodia of marine annelid worms, the ampullae and siphons of tunicates, and the tube feet of sea urchins. It was evident that the gene must be ancient, dating back to the last common ancestor of bilateral animals (before the Ediacaran Period, which began some 635 million years ago). Evo-devo had started to uncover the ways that all animal bodies were built during development.[30][31]
The control of body structure
Deep homology
Further information: Deep homologyRoughly spherical eggs of different animals give rise to extremely different bodies, from jellyfish to lobsters, butterflies to elephants. Many of these organisms share the same structural genes for body-building proteins like collagen and enzymes, but biologists had expected that each group of animals would have its own rules of development. The surprise of evo-devo is that the shaping of bodies is controlled by a rather small percentage of genes, and that these regulatory genes are ancient, shared by all animals. The giraffe does not have a gene for a long neck, any more than the elephant has a gene for a big body. Their bodies are patterned by a system of switching which causes development of different features to begin earlier or later, to occur in this or that part of the embryo, and to continue for more or less time.[7]
The puzzle of how embryonic development was controlled began to be solved using the fruit fly Drosophila melanogaster as a model organism. The step-by-step control of its embryogenesis was visualized by attaching fluorescent dyes of different colours to specific types of protein made by genes expressed in the embryo.[7] A dye such as green fluorescent protein, originally from a jellyfish, was typically attached to an antibody specific to a fruit fly protein, forming a precise indicator of where and when that protein appeared in the living embryo.[32]
Using such a technique, in 1994 Walter Gehring found that the pax-6 gene, vital for forming the eyes of fruit flies, exactly matches an eye-forming gene in mice and humans. The same gene was quickly found in many other groups of animals, such as squid, a cephalopod mollusc. Biologists including Ernst Mayr had believed that eyes had arisen in the animal kingdom at least 40 times, as the anatomy of different types of eye varies widely.[7] For example, the fruit fly's compound eye is made of hundreds of small lensed structures (ommatidia); the human eye has a blind spot where the optic nerve enters the eye, and the nerve fibres run over the surface of the retina, so light has to pass through a layer of nerve fibres before reaching the detector cells in the retina, so the structure is effectively "upside-down"; in contrast, the cephalopod eye has the retina, then a layer of nerve fibres, then the wall of the eye "the right way around".[33] The evidence of pax-6, however, was that the same genes controlled the development of the eyes of all these animals, suggesting that they all evolved from a common ancestor.[7] Ancient genes had been conserved through millions of years of evolution to create dissimilar structures for similar functions, demonstrating deep homology between structures once thought to be purely analogous.[34][35] This has caused a radical revision of the meaning of homology in evolutionary biology.[34][35][36]The pax-6 gene controls development of eyes of different types across the animal kingdom.
Gene toolkit
Expression of homeobox (Hox) genes in the fruit flyMain article: Evo-devo gene toolkitA small fraction of the genes in an organism's genome control the organism's development. These genes are called the developmental-genetic toolkit. They are highly conserved among phyla, meaning that they are ancient and very similar in widely separated groups of animals. Differences in deployment of toolkit genes affect the body plan and the number, identity, and pattern of body parts. Most toolkit genes are parts of signalling pathways: they encode transcription factors, cell adhesion proteins, cell surface receptor proteins and signalling ligands that bind to them, and secreted morphogens that diffuse through the embryo. All of these help to define the fate of undifferentiated cells in the embryo. Together, they generate the patterns in time and space which shape the embryo, and ultimately form the body plan of the organism. Among the most important toolkit genes are the Hox genes. These transcription factors contain the homeobox protein-binding DNA motif, also found in other toolkit genes, and create the basic pattern of the body along its front-to-back axis.[36] Hox genes determine where repeating parts, such as the many vertebrae of snakes, will grow in a developing embryo or larva.[7] Pax-6, already mentioned, is a classic toolkit gene.[37] Homeobox genes are also found in plants, implying they are common to all eukaryotes.[38][39][40]
The embryo's regulatory networks
Further information: Regulation of gene expression and Transcriptional regulationThe protein products of the regulatory toolkit are reused not by duplication and modification, but by a complex mosaic of pleiotropy, being applied unchanged in many independent developmental processes, giving pattern to many dissimilar body structures.[36] The loci of these pleiotropic toolkit genes have large, complicated and modular cis-regulatory elements. For example, while a non-pleiotropic rhodopsin gene in the fruit fly has a cis-regulatory element just a few hundred base pairs long, the pleiotropic eyeless cis-regulatory region contains 6 cis-regulatory elements in over 7000 base pairs.[36] The regulatory networks involved are often very large. Each regulatory protein controls "scores to hundreds" of cis-regulatory elements. For instance, 67 fruit fly transcription factors controlled on average 124 target genes each.[36] All this complexity enables genes involved in the development of the embryo to be switched on and off at exactly the right times and in exactly the right places. Some of these genes are structural, directly forming enzymes, tissues and organs of the embryo. But many others are themselves regulatory genes, so what is switched on is often a precisely-timed cascade of switching, involving turning on one developmental process after another in the developing embryo.[36]
Such a cascading regulatory network has been studied in detail in the development of the fruit fly embryo. The young embryo is oval in shape, like a rugby ball. A small number of genes produce messenger RNAs that set up concentration gradients along the long axis of the embryo. In the early embryo, the bicoid and hunchback genes are at high concentration near the anterior end, and give pattern to the future head and thorax; the caudal and nanos genes are at high concentration near the posterior end, and give pattern to the hindmost abdominal segments. The effects of these genes interact; for instance, the Bicoid protein blocks the translation of caudal's messenger RNA, so the Caudal protein concentration becomes low at the anterior end. Caudal later switches on genes which create the fly's hindmost segments, but only at the posterior end where it is most concentrated.[41][42]Gene product distributions along the long axis of the early embryo of a fruit fly
The Bicoid, Hunchback and Caudal proteins in turn regulate the transcription of gap genes such as giant, knirps, Krüppel, and tailless in a striped pattern, creating the first level of structures that will become segments.[26] The proteins from these in turn control the pair-rule genes, which in the next stage set up 7 bands across the embryo's long axis. Finally, the segment polarity genes such as engrailed split each of the 7 bands into two, creating 14 future segments.[41][42]
This process explains the accurate conservation of toolkit gene sequences, which has resulted in deep homology and functional equivalence of toolkit proteins in dissimilar animals (seen, for example, when a mouse protein controls fruit fly development). The interactions of transcription factors and cis-regulatory elements, or of signalling proteins and receptors, become locked in through multiple usages, making almost any mutation deleterious and hence eliminated by natural selection.[36]
The origins of novelty
Among the more surprising and, perhaps, counterintuitive (from a neo-Darwinian viewpoint) results of recent research in evolutionary developmental biology is that the diversity of body plans and morphology in organisms across many phyla are not necessarily reflected in diversity at the level of the sequences of genes, including those of the developmental genetic toolkit and other genes involved in development. Indeed, as Gerhart and Kirschner have noted, there is an apparent paradox: "where we most expect to find variation, we find conservation, a lack of change".[43] So, if the observed morphological novelty between different clades does not come from changes in gene sequences (such as by mutation), where does it come from? Novelty may arise by mutation-driven changes in gene regulation.[36][44][45][46]
Variations in the toolkit
Variations in the toolkit may have produced a large part of the morphological evolution of animals. The toolkit can drive evolution in two ways. A toolkit gene can be expressed in a different pattern, as when the beak of Darwin's large ground-finch was enlarged by the BMP gene,[47] or when snakes lost their legs as distal-less became under-expressed or not expressed at all in the places where other reptiles continued to form their limbs.[48] Or, a toolkit gene can acquire a new function, as seen in the many functions of that same gene, distal-less, which controls such diverse structures as the mandible in vertebrates,[49][50] legs and antennae in the fruit fly,[51] and eyespot pattern in butterfly wings.[52] Given that small changes in toolbox genes can cause significant changes in body structures, they have often enabled the same function convergently or in parallel. distal-less generates wing patterns in the butterflies Heliconius erato and Heliconius melpomene, which are Müllerian mimics. In so-called facilitated variation,[53] their wing patterns arose in different evolutionary events, but are controlled by the same genes.[54] Developmental changes can contribute directly to speciation.[55]Different species of Heliconius butterfly have independently evolved similar patterns, apparently both facilitated and constrained by the available developmental-genetic toolkit genes controlling wing pattern formation.
Consolidation of epigenetic changes
Main article: Transgenerational epigenetic inheritanceFurther information: Extended Evolutionary SynthesisEvolutionary innovation may sometimes begin in Lamarckian style with epigenetic alterations of gene regulation or phenotype generation, subsequently consolidated by changes at the gene level. Epigenetic changes include modification of DNA by reversible methylation,[56] as well as nonprogrammed remoulding of the organism by physical and other environmental effects due to the inherent plasticity of developmental mechanisms.[57] The biologists Stuart A. Newman and Gerd B. Müller have suggested that organisms early in the history of multicellular life were more susceptible to this second category of epigenetic determination than are modern organisms, providing a basis for early macroevolutionary changes.[58]
Eco-evo-devo
Ecological evolutionary developmental biology (eco-evo-devo) integrates research from developmental biology and ecology to examine their relationship with evolutionary theory.[59] Researchers study concepts and mechanisms such as developmental plasticity, epigenetic inheritance, genetic assimilation, niche construction and symbiosis.[60][61]
See also
- Cell signaling
- Evolution & Development (journal)
- Just So Stories (as seen by evolutionary developmental biologists)
Notes
- Richardson and Keuck (2002), "Haeckel's ABC of evolution and development", Biological reviews of the Cambridge Philosophical Society, 77 (4): 495–528, doi:10.1017/s1464793102005948, PMID 12475051 p.516
- Gilbert, S.F.; Bosch, T.C.G.; Ledón-Rettig, C. (2015). "Eco-Evo-Devo: developmental symbiosis and developmental plasticity as evolutionary agents". Nature Reviews Genetics. 16 (10): 611–622. doi:10.1038/nrg3982. PMID 26370902.
References
- Secord, James A. (2000), Victorian sensation: the extraordinary publication, reception, and secret authorship of Vestiges of the natural history of creation, Chicago: University of Chicago Press, ISBN 978-0-226-74410-0
Biologists could say, with confidence, that forms change, and that natural selection is an important force for change. Yet they could say nothing about how that change is accomplished. How bodies or body parts change, or how new structures arise, remained complete mysteries.
Cirripedes afford a good instance of this: even the illustrious Cuvier did not perceive that a barnacle was, as it certainly is, a crustacean; but a glance at the larva shows this to be the case in an unmistakeable manner.
Homeobox genes are found in almost all eukaryotes, and have diversified into 11 gene classes and over 100 gene families in animal evolution, and 10 to 14 gene classes in plants.









No comments:
Post a Comment